BioFormats
Store raw data from BioFormats images
Usage
OBJ = BioFormats(FILENAME, CHS, CAL)
Arguments
FILENAMEis the original filename (including the path) of thisBioFormatsobject.CHSis a scalar structure that contains information about the meaning of the image channels.CALis a scalarCalibrationPixelSizeobject.
Details
BioFormats objects are used to contain all raw image data and metadata from Bio-Formats Tif images. This class relies on existing code from the Bio-Formats library. For more information, please visit http://www.openmicroscopy.org/site/products/bio-formats.
See Also
RawImgclass documentationMetadataclass documentationMetadataquick start guideCalibrationPixelSizeclass documentationCalibrationPixelSizequick start guide
Examples
The following examples require the sample images and other files, which can be downloaded manually, from the University of Zurich website (http://www.pharma.uzh.ch/en/research/functionalimaging/CHIPS.html), or automatically, by running the function utils.download_example_imgs().
In addition, loading Bio-Formats images requires prior installation of the Bio-Formats Java library, which can be downloaded manually from the Open Microscopy Environment website (‘http://www.openmicroscopy.org/site/products/bio-formats), or automatically by running the function utils.install_bfmatlab().
Create a BioFormats object interactively
The following example will illustrate the process of creating a BioFormats object interactively. It is possible to do so by using the from_files() method, rather than the constructor (as is demonstrated in this section).
% Call the BioFormats constructor
bf001 = BioFormats()
Use the interactive dialogue box to select the raw image file cellscan_ome_tiff.ome.tif.
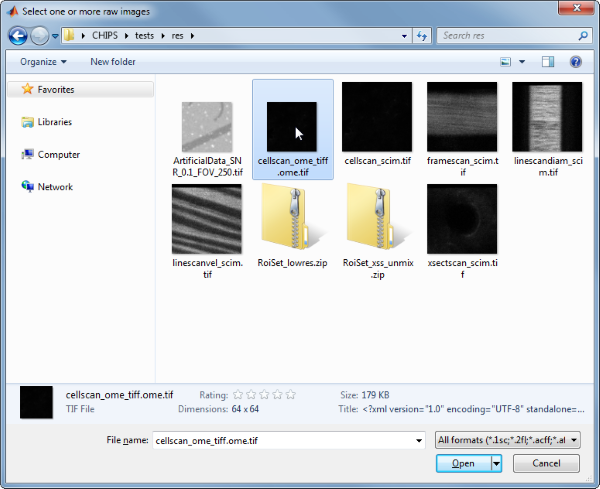
The next stage is to define the ‘meaning’ of the image channels. The first channel represents a cytosolic calcium sensor expressed in astrocytes. Press three and then enter to complete the selection.
----- What is shown on channel 1? -----
>> 0) <blank>
1) blood_plasma
2) blood_rbcs
3) Ca_Cyto_Astro
4) Ca_Memb_Astro
5) Ca_Neuron
6) cellular_signal
7) FRET_ratio
Answer: 3
We have now created a BioFormats object interactively.
bf001 =
BioFormats with properties:
filename: 'C:\...'
isDenoised: 0
isMotionCorrected: 0
metadata_original: [1x1 loci.formats.ome.OMEXMLMetadataImpl]
name: 'cellscan_ome_tiff.ome'
rawdata: [4-D uint16]
t0: 0
metadata: [1x1 Metadata]
The process is almost exactly the same to create an array of BioFormats objects; when the software prompts you to select one or more raw images, simply select multiple images by using either the shift or control key.
Create a BioFormats object without any interaction
% Specify the full path to the raw image object
fnST002 = fullfile(utils.CHIPS_rootdir, 'tests', 'res', ...
'cellscan_ome_tiff.ome.tif');
% Specify the channels relevant for this raw image
channels = struct('Ca_Cyto_Astro', 1);
% Create the SCIM_Tif object without any interaction
bf002 = BioFormats(fnST002, channels);
% View the SCIM_Tif object
bf002.plot()
Reading series #1
....................