Metadata
Store image metadata
Usage
OBJ = Metadata(IMGSIZE, ACQ, CHS, CAL)
Arguments
IMGSIZEis a vector of the dimensions of the raw image data.ACQis a scalar structure containing information about the image acquisition (e.g. line time, zoom factor etc).CHSis a scalar structure that contains information about the meaning of the image channels.CALis a scalarCalibrationPixelSizeobject.
Details
Metadata objects are used to contain all the extra information (i.e. metadata) about a given raw image. It also implements a number of related helper/convenience functions.
See Also
Metadataclass documentationCalibrationPixelSizeclass documentationCalibrationPixelSizequick start guideRawImgclass documentationRawImgquick start guide
Examples
The following examples require the sample images and other files, which can be downloaded manually, from the University of Zurich website (http://www.pharma.uzh.ch/en/research/functionalimaging/CHIPS.html), or automatically, by running the function utils.download_example_imgs().
Create a Metadata object interactively
The following example will illustrate the process of creating a Metadata object interactively. Normally this process is done automatically when creating a RawImg object, but in certain circumstances it is useful to do it manually.
% Call the Metadata constructor
md001 = Metadata()
Answer some questions about the image acquisition. For example, specify that the image was not bidirectional, that the line time was 2ms, and that the zoom factor was 4.
Please enter a value for if the image is bidirectional [1/0]: 0
Please enter a value for the line time [ms]: 2
Please enter a value for the zoom factor: 4
Use the interactive dialogue box to select the dummy calibration (calibration_dummy.mat), which should be located in the subfolder tests>res, within the CHIPS root directory:
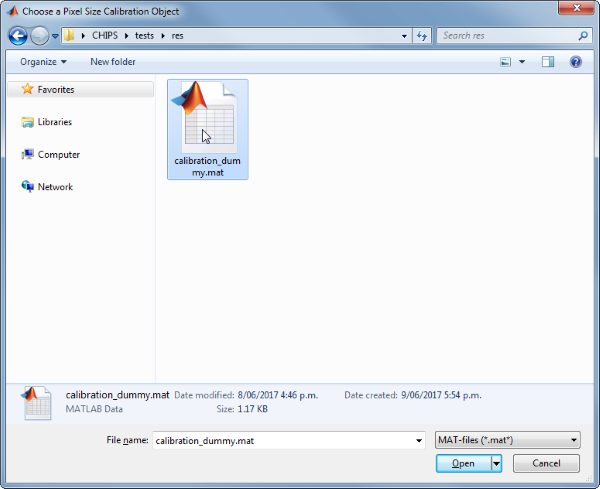
We have now created a Metadata object interactively.
md001 =
Warning: The original number of lines per frame is not defined for this image. This may be because the metadata was created in an unexpected way. Please check carefully any results that depend on the frame rate. > In Metadata/get.frameRate (line 433) Warning: The image dimensions are not defined, so the frame rate could not be determined. > In Metadata/get.frameRate (line 444) Warning: The original number of pixels per line is not defined for this image. This may be because the metadata was created in an unexpected way. Please check carefully any results that depend on the pixel size. > In Metadata/get.pixelSize (line 469) Warning: The image dimensions are not defined, so the pixel size could not be determined. > In Metadata/get.pixelSize (line 480)
Metadata with properties:
calibration: [1x1 CalibrationPixelSize]
channels: []
discardFlybackLine: []
frameRate: NaN
isBiDi: 0
lineTime: 2
nChannels: []
nFrames: []
nLinesPerFrame: []
nLinesPerFrameOrig: []
nPixelsPerLine: []
nPixelsPerLineOrig: []
pixelSize: NaN
pixelTime: []
zoom: 4
knownChannels: {1x7 cell}
Create a Metadata object without any interaction
% Specify the image dimensions
imgSize = [128, 128, 2, 10];
% Specify some data about the image acquisition
acq = struct('isBiDi', false, 'lineTime', 2, 'zoom', 4, ...
'nLinesPerFrameOrig', imgSize(1), 'nPixelsPerLineOrig', imgSize(2));
% Specify the channels relevant for this raw image
channels = struct('Ca_Cyto_Astro', 1, 'blood_plasma', 2);
% Load the CalibrationPixelSize object
fnCalibration = fullfile(utils.CHIPS_rootdir, 'tests', 'res', ...
'calibration_dummy.mat');
calibration = CalibrationPixelSize.load(fnCalibration);
% Create the Metadata object without any interaction
md002 = Metadata(imgSize, acq, channels, calibration)
md002 =
Metadata with properties:
calibration: [1×1 CalibrationPixelSize]
channels: [1×1 struct]
discardFlybackLine: []
frameRate: 3.9063
isBiDi: 0
lineTime: 2
nChannels: 2
nFrames: 10
nLinesPerFrame: 128
nLinesPerFrameOrig: 128
nPixelsPerLine: 128
nPixelsPerLineOrig: 128
pixelSize: 2
pixelTime: []
zoom: 4
knownChannels: {1×7 cell}